What is a column constant?
When you fit a number of datasets at once, you can use the column title as a second independent variable. We call this constraining a parameter to be a column constant. This is best seen by example.
How to enter column constants
To see how column constants work, create a new XY table using the sample data file: Enzyme kinetics - Competitive inhibition.
The data table has one X column, and four Y columns, each representing a different concentration of inhibitor. The inhibitor concentrations are entered as column title.
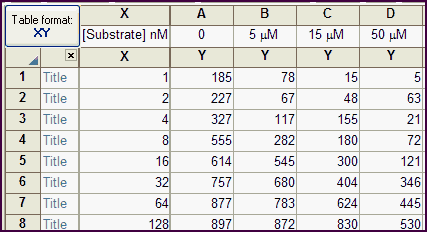
Note that Prism reads only the number in the column title. In this example, the units are specified as micromolar, but Prism ignores this and simply reads the numbers. It does not do any unit conversion.
Specifying a column constant when fitting data
When choosing an initial value, choose from the drop down list either "Mean of column title value" or "log(Mean of column title value".

Example
To fit the sample data above, click Analyze, choose nonlinear regression, choose the Enzyme Kinetics panel of equations and choose Competitive enzyme kinetics. The equation is built in, but if you click the Details button you can see the math.
KmObs=Km(1+[I]/Ki)
Y=Vmax*X/(KmObs+X)
The first line defines an intermediate variable (KmObs, the observed Michaelis-Menten constant in the presence of a competitive inhibitor), which is a function of the Michaelis-Menten constant of the enzyme (Km), the concentration of inhibitor (I), and the competitive inhibition constant (Ki).
The second line computes enzyme velocity (Y) as a function of substrate concentration (X) and KMapp.
This model is defined with I constrained to being a data set constant, which means its value comes from the column titles. In this example, therefore, I=0 when fitting column A, I=5 when fitting column B, etc. The 'μM' in the title is ignored by Prism -- it doesn't do any unit conversions.
The other three parameters (Km, Ki and Vmax) are defined to be shared, so Prism fits one best-fit value that applies to the entire family of datasets.

Prism determined the maximum velocity of the enzyme with no inhibitor (Vmax in the same units as the Y values you entered), the Michaelis-Menten constant of the enzyme with no inhibitor (Km, in the units used for X values) and the competitive inhibition constant (Ki, in units used for the column constants). Note that I is not a parameter to be fit, but rather takes on constant values you entered into the column titles. KmObs is not a parameter to be fit, but is rather an intermediate variable used to define the model.
Learn more about competitive enzyme inhibition.
Summary. The advantage of column constants
By using column constants and global fitting (shared parameters), this example determined a parameter (Ki) whose value cannot be determined from any one dataset, but can only be determined by examining the relationships between datasets.